Enzymes and receptors thermostability and activity
Many biotechnologically important enzymes show low thermal stability. The thermostability of enzymes is crucial in exoenergetic reactions, where a lot of heat can be released and thus enzyme can lose activity. Sometimes bioreactors are specially heated to prevent the multiplication of unwanted microorganisms. In some cases of industrial enzymes activity is low or is too narrow or too wide toward the spectrum of substrates.
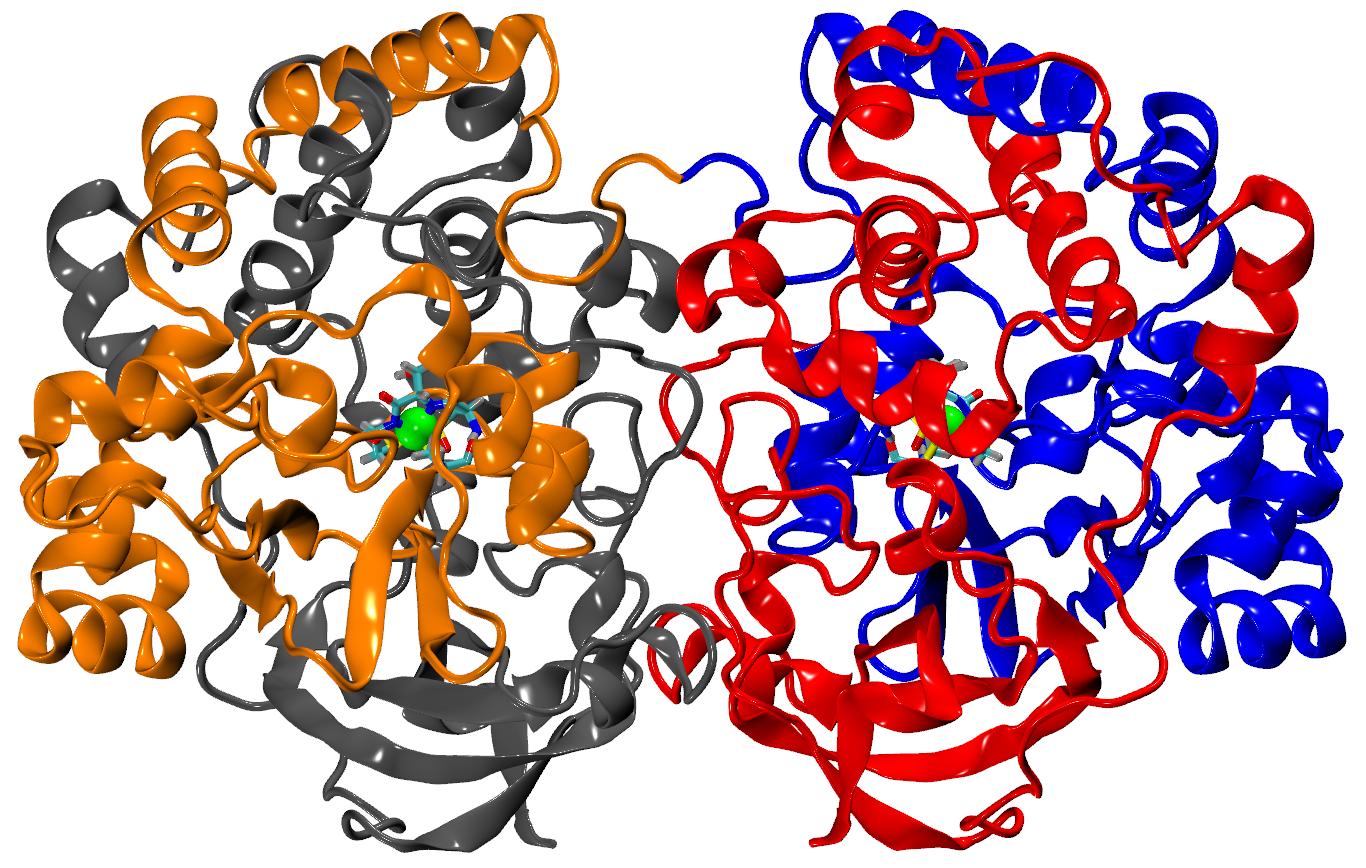 Figure 1. Nitrile hydratase, its nonstandard active site and reaction catalyzed by NHase.
Figure 1. Nitrile hydratase, its nonstandard active site and reaction catalyzed by NHase.
Nitrile hydratase (NHase, EC 4.2.1.84, Figure 1) is one of the most successful examples of using microbial enzyme in bioconversion of toxic nitriles to useful amides (Figure 2). This metalloprotein with posttranslationally modified cysteines is used in green chemistry in the production of many amides on the kiloton scale. Yearly over 600 kilotons of acrylamide and nicotinamide is produced using the NHase enzyme. Unfortunately wild types of NHase show low thermal stability and during large-scale bioconversion heated bioreactor destroys enzymes. In our group we are trying to explain why some mutational variants are more stable than others and to design new variants with improved thermostability [1, 2]. We also are trying to explain how the process of biotransformation of nitriles to amides occurs in NHase [3-5], how to re-design NHase to force this enzyme to be stereoselective [6] or regioselective [7], and how this enzyme is activated by metalochaperones [8]. We are using molecular modeling tools, like molecular dynamics simulations, docking studies, quantum calculations or bioinformatics. All studies are conducted in close cooperation with the experimental group of Zhemin Zhou from the School of Biotechnology, Jiangnan University, Wuxi, China.
- Cheng, Z.; Lan, Y.; Guo, J.; Ma, D.; Jiang, S.; Lai, Q.; Zhou, Z.; Peplowski, L., Computational Design of Nitrile Hydratase from Pseudonocardia thermophila JCM3095 for Improved Thermostability. Molecules 2020, 25, (20), 4806.
- Xia, Y.; Cui, W.; Cheng, Z.; Peplowski, L.; Liu, Z.; Kobayashi, M.; Zhou, Z., Improving the Thermostability and Catalytic Efficiency of the Subunit-Fused Nitrile Hydratase by Semi-Rational Engineering. ChemCatChem 2018, 10, (6), 1370-1375.
- Peplowski, L.; Kubiak, K.; Nowak, W., A Comparative DFT Study of Substrates and Products of Industral Enzyme Nitrile Hydratase Int J Quant Chem 2008, 108, 161-179.
- Peplowski, L.; Kubiak, K.; Nowak, W., Mechanical aspects of nitrile hydratase enzymatic activity. Steered molecular dynamics simulations of Pseudonocardia thermophila JCM 3095. Chem. Phys. Lett. 2008, 467, (1), 144-149.
- Peplowski, L.; Kubiak, K.; Nowak, W., Insights into catalytic activity of industrial enzyme Co-nitrile hydratase. Docking studies of nitriles and amides. J. Mol. Model. 2007, 13, (6-7), 725-30.
- Cheng, Z.; Peplowski, L.; Cui, W.; Xia, Y.; Liu, Z.; Zhang, J.; Kobayashi, M.; Zhou, Z., Identification of key residues modulating the stereoselectivity of nitrile hydratase toward rac-mandelonitrile by semi-rational engineering. Biotechnol. Bioeng. 2018, 115, (3), 524-535.
- Cheng, Z.; Cui, W.; Xia, Y.; Peplowski, L.; Kobayashi, M.; Zhou, Z., Modulation of Nitrile Hydratase Regioselectivity towards Dinitriles by Tailoring the Substrate Binding Pocket Residues. ChemCatChem 2018, 10, (2), 449-458.
- Xia, Y.; Peplowski, L.; Cheng, Z.; Wang, T.; Liu, Z.; Cui, W.; Kobayashi, M.; Zhou, Z., Metallochaperone function of the self-subunit swapping chaperone involved in the maturation of subunit-fused cobalt-type nitrile hydratase. Biotechnol. Bioeng. 2019, 116, (3), 481-489.