Ferroptosis and programmed cell death
Ferroptosis is a recently discovered form of cell death, different from better known apoptosis or necrosis. Ferroptosis characteristic feature is increasing level of lipid peroxides where free radicals take electrons from the lipids causing the cell damage. Ferroptosis was identified as the mechanism of cell death in Parkinson and Huntington’s diseases, and sepsis. It plays a critical role in the treatment of cancers, and may contribute to the degradation of tissue in brain trauma, kidney diseases and asthma.
Dr. Karolina Mikulska-Ruminska investigates with US collaborators the process of ferroptosis. Explanation of ferroptosis at the molecular level using computational methods and its experimental verification is essential for developing new drugs and treatment methods. We are using a wide range of computational and bioinformatics methods such as all-atom and coarse-grained molecular dynamics, molecular docking, homology modeling, elastic network models (GNM, ANM), phylogenetic analysis and more to better understand this process.
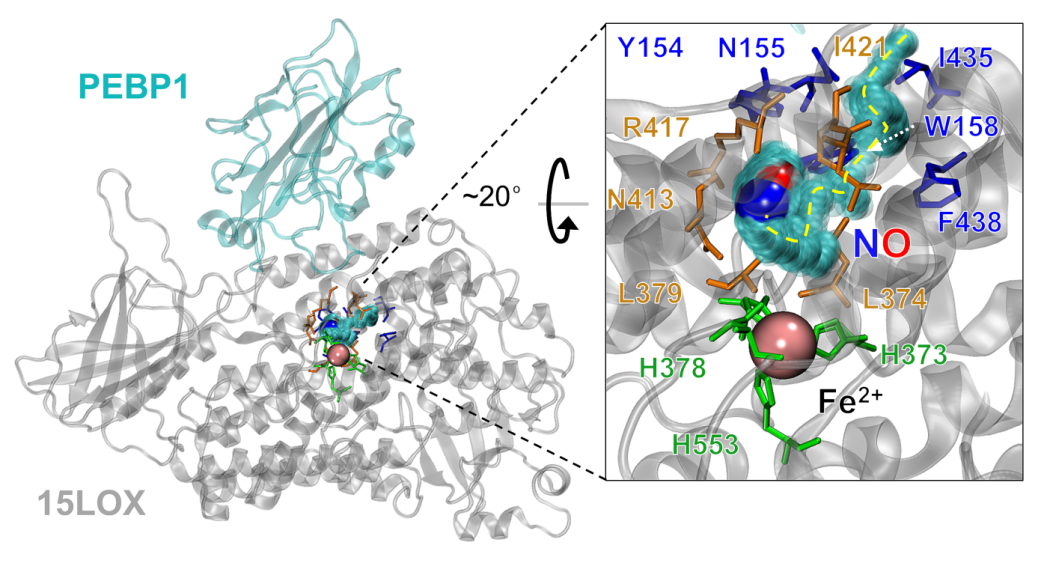
Figure 1. Computational model of PEBP1-lypoxygenase complex (PEBP1-15LOX, coordinates for both proteins obtained from X-ray) with catalytic site which consist of iron ion (denoted as the pink sphere) that is covalently bonded to histidines and isoleucine from lipoxygenase (green residues). Residues in blue belong to the entrance of the tunnel localized in the lipoxygenase. As a result of the interactions with nitric oxide the tunnel is opening. Residues that stabilize the interactions with nitric oxide in the catalytic site are displayed in orange. Yellow, dashed line indicates the nitric oxide pathway in lipoxygenase structure.
- S. E. Wenzel, Y. Y. Tyurina, J. Zhao, C. M. S. Croix, H. H. Dar, G. Mao, V. A. Tyurin, T. S. Anthonymuthu, A. A. Kapralov, A. A. Amoscato, K. Mikulska-Ruminska, I. H. Shrivastava, E. M. Kenny, Q. Yang, J. C. Rosenbaum, L. J. Sparvero, D. R. Emlet, X. Wen, Y. Minami, F. Qu, S. C. Watkins, T. R. Holman, A. P. VanDemark, J. A. Kellum, I. Bahar, H. Bayır, and V. E. Kagan
Cell 171, 628-641. e626 (2017). - T. S. Anthonymuthu, Y. Y. Tyurina, W.-Y. Sun, K. Mikulska-Ruminska, I. H. Shrivastava, V. A. Tyurin, F. B. Cinemre, H. H. Dar, A. P. VanDemark, T. R. Holman, Y. Sadovsky, B. R. Stockwell, R.-R. He, I. Bahar, H. Bayır, and V. E. Kagan
Redox Biology, 101744 (2020). - A. A. Kapralov, Q. Yang, H. H. Dar, Y. Y. Tyurina, T. S. Anthonymuthu, R. Kim, C. M. S. Croix, K. Mikulska-Ruminska, B. Liu, I. H. Shrivastava, V. A. Tyurin, H.-C. Ting, Y. L. Wu, Y. Gao, G. V. Shurin, M. A. Artyukhova, L. A. Ponomareva, P. S. Timashev, R. M. Domingues, D. A. Stoyanovsky, J. S. Greenberger, R. K. Mallampalli, I. Bahar, D. I. Gabrilovich, H. Bayır, and V. E. Kagan
Nature Chemical Biology 16, 278-290 (2020). - K. Mikulska-Ruminska, I. Shrivastava, J. Krieger, S. Zhang, H. Li, H. Bayır, S. E. Wenzel, A. P. VanDemark, V. E. Kagan, and I. Bahar
J. Chem. Inform. Model. 59, 2496-2508 (2019). - T. S. Anthonymuthu, E. M. Kenny, I. Shrivastava, Y. Y. Tyurina, Z. E. Hier, H.-C. Ting, H. H. Dar, V. A. Tyurin, A. Nesterova, A. A. Amoscato, K. Mikulska-Ruminska, J. C. Rosenbaum, G. Mao, J. Zhao, M. Conrad, J. A. Kellum, S. E. Wenzel, A. P. VanDemark, I. Bahar, V. E. Kagan, and H. l. Bayir
J. Am. Chem. Soc. 140, 17835-17839 (2018). - H. H. Dar, Y. Y. Tyurina, K. Mikulska-Ruminska, I. Shrivastava, H.-C. Ting, V. A. Tyurin, J. Krieger, C. M. St. Croix, S. Watkins, E. Bayir, G. Mao, C. R. Armbruster, A. Kapralov, H. Wang, M. R. Parsek, T. S. Anthonymuthu, A. F. Ogunsola, B. A. Flitter, C. J. Freedman, J. R. Gaston, T. R. Holman, J. M. Pilewski, J. S. Greenberger, R. K. Mallampalli, Y. Doi, J. S. Lee, I. Bahar, J. M. Bomberger, H. Bayır, and V. E. Kagan
J. Clin. Invest. 128, 4639-4653 (2018).